Generalizing biomedical relation classification with neural adversarial domain adaptation
Anthony Rios, Ramakanth Kavuluru, and Zhiyong Lu
Bioinformatics, Volume 34, Issue 17, 1 September 2018, Pages 2973–2981
Abstract:
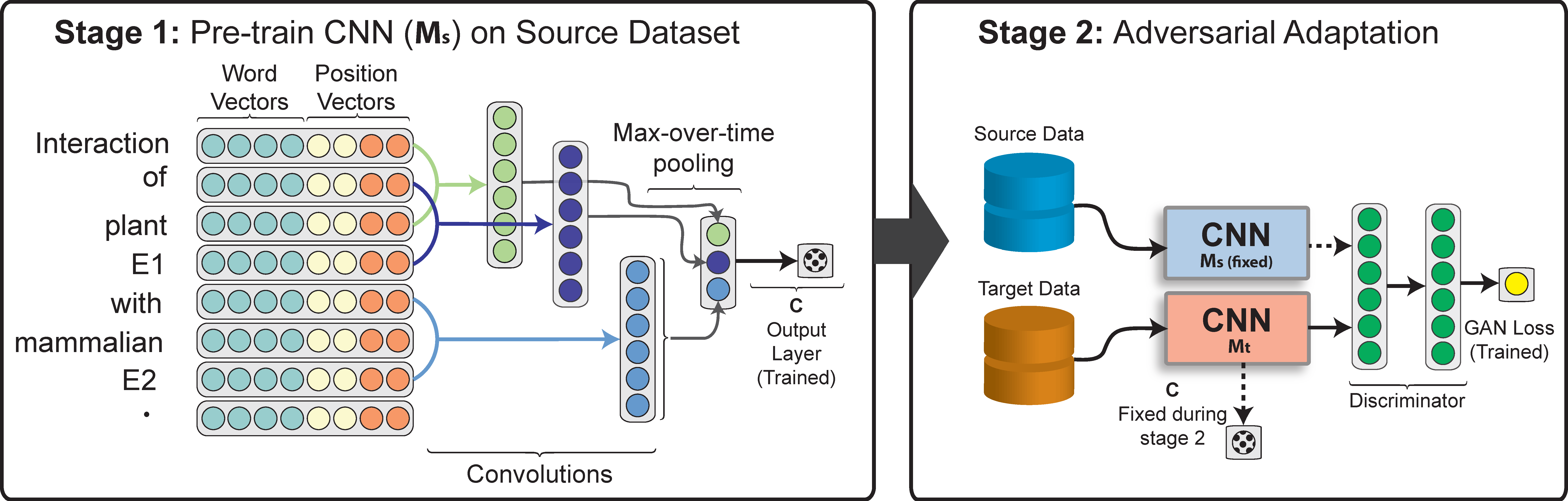
Creating large datasets for biomedical relation classification can be prohibitively expen- sive. While some datasets have been curated to extract protein–protein and drug–drug interactions (PPIs and DDIs) from text, we are also interested in other interactions including gene–disease and chemical–protein connections. Also, many biomedical researchers have begun to explore ternary relationships. Even when annotated data are available, many datasets used for relation classification are inherently biased. For example, issues such as sample selection bias typically prevent models from generalizing in the wild. To address the problem of cross-corpora generalization, we present a novel adversarial learning algorithm for unsupervised domain adaptation tasks where no labeled data are available in the target domain. Instead, our method takes advantage of unlabeled data to improve biased classifiers through learning domain-invariant features via an adversarial process. Finally, our method is built upon recent advances in neural network (NN) methods.
Results: We experiment by extracting PPIs and DDIs from text. In our experiments, we show domain invariant features can be learned in NNs such that classifiers trained for one interaction type (protein–protein) can be re-purposed to others (drug–drug). We also show that our method can adapt to different source and target pairs of PPI datasets. Compared to prior convolutional and recurrent NN-based relation classification methods without domain adaptation, we achieve improvements as high as 30% in F1-score. Likewise, we show improvements over state-of-the-art adversarial methods.
@article{doi:10.1093/bioinformatics/bty190,
author = {Rios, Anthony and Kavuluru, Ramakanth and Lu, Zhiyong},
title = {Generalizing biomedical relation classification with neural adversarial domain adaptation},
journal = {Bioinformatics},
volume = {34},
number = {17},
pages = {2973-2981},
year = {2018},
doi = {10.1093/bioinformatics/bty190},
URL = {http://dx.doi.org/10.1093/bioinformatics/bty190},
eprint = {/oup/backfile/content_public/journal/bioinformatics/34/17/10.1093_bioinformatics_bty190/2/bty190.pdf}
}